|
PaGMO
1.1.5
|
|
PaGMO
1.1.5
|
Monotonic Basin Hopping (generalized) More...
#include <mbh.h>
Public Member Functions | |
| mbh (const base &=cs(), int stop=5, double perturb=5e-2) | |
| Constructor. More... | |
| mbh (const base &, int stop, const std::vector< double > &perturb) | |
| Constructor. More... | |
| mbh (const mbh &) | |
| Copy constructor. | |
| base_ptr | clone () const |
| Clone method. | |
| void | evolve (population &) const |
| Evolve implementation. More... | |
| std::string | get_name () const |
| Algorithm name. | |
| base_ptr | get_algorithm () const |
| Get a copy of the internal local algorithm. More... | |
| void | set_algorithm (const base &) |
| Set algorithm. More... | |
 Public Member Functions inherited from pagmo::algorithm::base Public Member Functions inherited from pagmo::algorithm::base | |
| base () | |
| Default constructor. More... | |
| virtual | ~base () |
| Trivial destructor. More... | |
| std::string | human_readable () const |
| Return human readable representation of the algorithm. More... | |
| void | set_screen_output (const bool p) |
| Setter-Getter for protected m_screen_output data. More... | |
| bool | get_screen_output () const |
| Gets screen output. More... | |
| void | reset_rngs (const unsigned int) const |
| Resets the seed of the internal rngs using a user-provided seed. More... | |
Protected Member Functions | |
| std::string | human_readable_extra () const |
| Extra human readable algorithm info. More... | |
Friends | |
| class | boost::serialization::access |
Additional Inherited Members | |
 Protected Attributes inherited from pagmo::algorithm::base Protected Attributes inherited from pagmo::algorithm::base | |
| bool | m_screen_output |
| Indicates to the derived class whether to print stuff on screen. | |
| rng_double | m_drng |
| Random number generator for double-precision floating point values. | |
| rng_uint32 | m_urng |
| Random number generator for unsigned integer values. | |
| unsigned int | m_fevals |
| A counter for the number of function evaluations. | |
Monotonic Basin Hopping (generalized)
Monotonic basin hopping, or simply, basin hopping, is an algorithm rooted in the idea of transforming each the objective function at  into the local minima found starting from
into the local minima found starting from 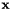 This simple idea allowed a substantial increase of efficiency in solving problems, such as the Lennard-Jones cluster or the MGA-DSM interplanetary trajectory problem that are conjectured to have a so-called funnel structure.
This simple idea allowed a substantial increase of efficiency in solving problems, such as the Lennard-Jones cluster or the MGA-DSM interplanetary trajectory problem that are conjectured to have a so-called funnel structure.
In PaGMO we provide an original generalization of the method that operates on any pagmo::population using any pagmo::algorithm. When a population containing a single individual is used and coupled with a local optimizer, the original method is recovered. The pseudo code of the generalized version is:
> Select a pagmo::population > Select a pagmo::algorithm > Store best individual > while i < stop_criteria > > Perturb the population in a selected neighbourhood > > Evolve the population using the algorithm > > if the best individual is improved (according to the problem::compare_fc criteria) > > > increment i > > > update best individual > > else > > > i = 0
Constructor.
Constructs an mbh algorithm with uniform neighbourhoods
| [in] | local | pagmo::algorithm to use as 'local' optimization method |
| [in] | stop | number of consecutive step allowed without any improvement |
| [in] | perturb | At the end of one iteration of mbh, each chromosome of each individual will be perturbed within +-perturb*(ub-lb), the same for the velocity. The integer part is treated the same way. rounding to the floor |
| value_error | if stop is negative or perturb is not in [0,1] |
| pagmo::algorithm::mbh::mbh | ( | const base & | local, |
| int | stop, | ||
| const std::vector< double > & | perturb | ||
| ) |
Constructor.
Allows to specify in detail all the parameters of the algorithm.
| [in] | local | pagmo::algorithm to use as 'local' optimization method |
| [in] | stop | number of consecutive step allowed without any improvement |
| [in] | perturb | At the end of one iteration of mbh, the i-th chromosome of each individual will be perturbed within +-perturb[i]*(ub[i]-lb[i]), the same for the velocity. The integer part is treated the same way rounding to the floor |
| value_error | if stop is negative or perturb[i] is not in [0,1] |
|
virtual |
Evolve implementation.
Run the MBH algorithm
| [in,out] | pop | input/output pagmo::population to be evolved. |
Implements pagmo::algorithm::base.
| base_ptr pagmo::algorithm::mbh::get_algorithm | ( | ) | const |
Get a copy of the internal local algorithm.
|
protectedvirtual |
Extra human readable algorithm info.
Reimplemented from pagmo::algorithm::base.
| void pagmo::algorithm::mbh::set_algorithm | ( | const base & | algo | ) |
 1.8.9.1
1.8.9.1