Point to point Sims-FLanagan#
In this tutorial we show the use of the pykep.trajopt.sf_point2point to find a low-thrust trajectory connecting two fixed points in space.
Since the points are considered fixed this effort has mainly an academic value, but it is infomrative in the study of the numerical properties of an optimization pipeline.
The decision vector in this class compatible with pygmo [BI20] UDPs (User Defined Problems) is:
\[
\mathbf x = [m_f, u_{x0}, u_{y0}, u_{z0}, u_{x1}, u_{y1}, u_{z1}, ..., T_{tof}]
\]
Note
This notebook makes use of the commercial solver SNOPT 7 and to run needs a valid snopt_7_c library installed in the system. In case SNOPT7 is not available, you can still run the notebook using, for example uda = pg.algorithm.nlopt("slsqp") with minor modifications.
Basic imports:
import pykep as pk
import numpy as np
import time
import pygmo as pg
import pygmo_plugins_nonfree as ppnf
import time
from matplotlib import pyplot as plt
# Problem data
mu = pk.MU_SUN
max_thrust = 0.22
isp = 3000
# Initial state
ms = 1000.0
rs = np.array([1.2, 0.0, -0.01]) * pk.AU
vs = np.array([0.01, 1, -0.01]) * pk.EARTH_VELOCITY
# Final state
rf = np.array([1, 0.0, -0.0]) * pk.AU
vf = np.array([0.01, 1.1, -0.0]) * pk.EARTH_VELOCITY
# Throttles and tof
nseg = 4
throttles = np.random.uniform(-1, 1, size=(nseg * 3))
tof = 2 * np.pi * np.sqrt(pk.AU**3 / pk.MU_SUN) / 4
udp_nog = pk.trajopt.sf_point2point(
rvs=[rs, vs],
rvf=[rf, vf],
ms = 1000,
mu=pk.MU_SUN,
max_thrust=0.22,
veff=3000*pk.G0,
tof_bounds=[200, 500],
mf_bounds=[200.0, 1000.0],
nseg=nseg,
cut=0.6,
with_gradient=False,
)
udp_g = pk.trajopt.sf_point2point(
rvs=[rs, vs],
rvf=[rf, vf],
ms = 1000,
mu=pk.MU_SUN,
max_thrust=0.22,
veff=3000*pk.G0,
tof_bounds=[200, 500],
mf_bounds=[200.0, 1000.0],
nseg=nseg,
cut=0.6,
with_gradient=True,
)
snopt72 = "/Users/dario.izzo/opt/libsnopt7_c.dylib"
uda = ppnf.snopt7(library=snopt72, minor_version=2, screen_output=False)
uda.set_integer_option("Major iterations limit", 2000)
uda.set_integer_option("Iterations limit", 20000)
uda.set_numeric_option("Major optimality tolerance", 1e-3)
uda.set_numeric_option("Major feasibility tolerance", 1e-12)
# uda = pg.nlopt("slsqp")
algo = pg.algorithm(uda)
prob_nog = pg.problem(udp_nog)
prob_nog.c_tol = 1e-6
prob_g = pg.problem(udp_g)
prob_g.c_tol = 1e-6
pop_g = pg.population(prob_g, 1)
pop_g = algo.evolve(pop_g)
print(prob_g.feasibility_f(pop_g.champion_f))
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[4], line 6
4 prob_g.c_tol = 1e-6
5 pop_g = pg.population(prob_g, 1)
----> 6 pop_g = algo.evolve(pop_g)
7 print(prob_g.feasibility_f(pop_g.champion_f))
ValueError:
function: evolve_version
where: /home/conda/feedstock_root/build_artifacts/pygmo_plugins_nonfree_1761586710010/work/src/snopt7.cpp, 603
what:
An error occurred while loading the snopt7_c library at run-time. This is typically caused by one of the following
reasons:
- The file declared to be the snopt7_c library, i.e. /Users/dario.izzo/opt/libsnopt7_c.dylib, is not a shared library containing the necessary C interface symbols (is the file path really pointing to
a valid shared library?)
- The library is found and it does contain the C interface symbols, but it needs linking to some additional libraries that are not found
at run-time.
We report the exact text of the original exception thrown:
function: evolve_version
where: /home/conda/feedstock_root/build_artifacts/pygmo_plugins_nonfree_1761586710010/work/src/snopt7.cpp, 553
what: The snopt7_c library path was constructed to be: /Users/dario.izzo/opt/libsnopt7_c.dylib and it does not appear to be a file
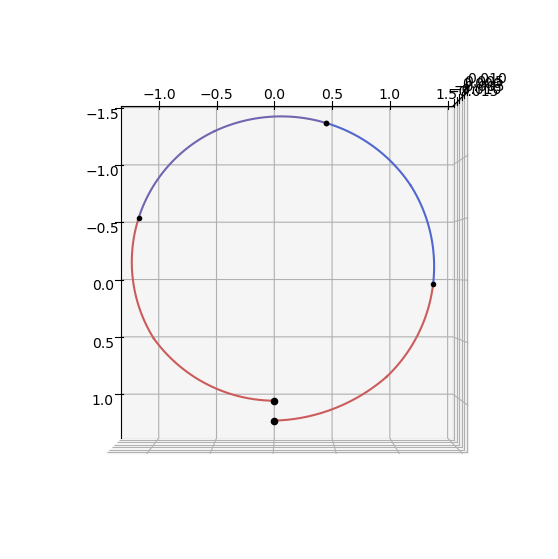
ax = udp_nog.plot(pop_g.get_x()[0], show_gridpoints=True)
ax.view_init(90, 0)
from tqdm import tqdm
cpu_nog = []
cpu_g = []
fail_g = 0
fail_nog = 0
for i in tqdm(range(2000)):
pop_nog = pg.population(prob_nog, 1)
pop_g = pg.population(prob_g)
pop_g.push_back(pop_nog.get_x()[0])
start = time.time()
pop_g = algo.evolve(pop_g)
end = time.time()
cpu_g.append(end - start)
if not prob_g.feasibility_f(pop_g.champion_f):
fail_g += 1
start = time.time()
pop_nog = algo.evolve(pop_nog)
end = time.time()
cpu_nog.append(end - start)
if not prob_nog.feasibility_f(pop_nog.champion_f):
fail_nog += 1
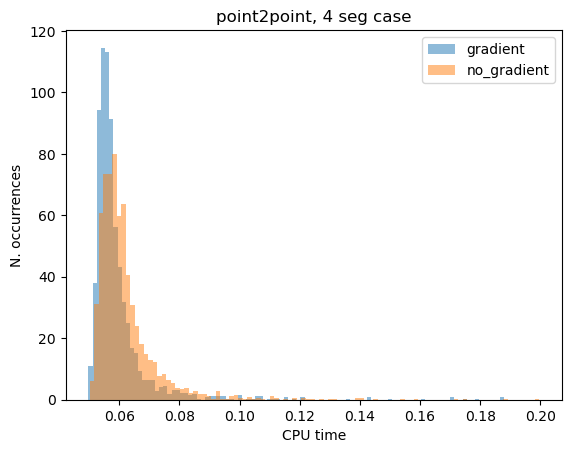
print(f"Gradient: {np.median(cpu_g):.4e}s")
print(f"No Gradient: {np.median(cpu_nog):.4e}s")
print(f"\nGradient (n.fails): {fail_g}")
print(f"No Gradient (n.fails): {fail_nog}")
100%|██████████| 2000/2000 [1:57:55<00:00, 3.54s/it]
Gradient: 2.7463e-01s
No Gradient: 2.7796e-01s
Gradient (n.fails): 192
No Gradient (n.fails): 189
cpu_g = np.array(cpu_g)
cpu_nog = np.array(cpu_nog)
plt.hist(cpu_g[cpu_g < 0.2], bins=100, label="gradient", density=True, alpha=0.5)
plt.hist(cpu_nog[cpu_nog < 0.2], bins=100, label="no_gradient", density=True, alpha=0.5)
# plt.xlim([0,0.15])
plt.legend()
plt.title("point2point, 4 seg case")
plt.xlabel("CPU time")
plt.ylabel("N. occurrences")
Text(0, 0.5, 'N. occurrences')