|
PaGMO
1.1.5
|
|
PaGMO
1.1.5
|
Nondominated Sorting genetic algorithm II (NSGA-II) More...
#include <nsga2.h>
Public Member Functions | |
| nsga2 (int gen=100, double cr=0.95, double eta_c=10, double m=0.01, double eta_m=50) | |
| Constructor. More... | |
| base_ptr | clone () const |
| Clone method. | |
| void | evolve (population &) const |
| Evolve implementation. More... | |
| std::string | get_name () const |
| Algorithm name. | |
 Public Member Functions inherited from pagmo::algorithm::base Public Member Functions inherited from pagmo::algorithm::base | |
| base () | |
| Default constructor. More... | |
| virtual | ~base () |
| Trivial destructor. More... | |
| std::string | human_readable () const |
| Return human readable representation of the algorithm. More... | |
| void | set_screen_output (const bool p) |
| Setter-Getter for protected m_screen_output data. More... | |
| bool | get_screen_output () const |
| Gets screen output. More... | |
| void | reset_rngs (const unsigned int) const |
| Resets the seed of the internal rngs using a user-provided seed. More... | |
Protected Member Functions | |
| std::string | human_readable_extra () const |
| Extra human readable algorithm info. More... | |
Friends | |
| class | boost::serialization::access |
Additional Inherited Members | |
 Protected Attributes inherited from pagmo::algorithm::base Protected Attributes inherited from pagmo::algorithm::base | |
| bool | m_screen_output |
| Indicates to the derived class whether to print stuff on screen. | |
| rng_double | m_drng |
| Random number generator for double-precision floating point values. | |
| rng_uint32 | m_urng |
| Random number generator for unsigned integer values. | |
| unsigned int | m_fevals |
| A counter for the number of function evaluations. | |
Nondominated Sorting genetic algorithm II (NSGA-II)
NSGA-II is a nondominated-sorting based multiobjective evolutionary algorithm. It genererates offspring with crossover and mutation and select the next generation according to nondominated-sorting and crowding distance comparison.
The algorithm can be applied to continuous box-bounded optimization. The version for mixed integer and constrained optimization is also planned.
| pagmo::algorithm::nsga2::nsga2 | ( | int | gen = 100, |
| double | cr = 0.95, |
||
| double | eta_c = 10, |
||
| double | m = 0.01, |
||
| double | eta_m = 50 |
||
| ) |
Constructor.
Constructs a NSGA II algorithm
| [in] | gen | Number of generations to evolve. |
| [in] | cr | Crossover probability |
| [in] | eta_c | Distribution index for crossover |
| [in] | m | Mutation probability |
| [in] | eta_m | Distribution index for mutation |
| value_error | if gen is negative, crossover probability is not 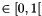 , mutation probability or mutation width is not , mutation probability or mutation width is not ![$ \in [0,1]$](form_6.png) , , |
|
virtual |
Evolve implementation.
Run the NSGA-II algorithm for the number of generations specified in the constructors.
| [in,out] | pop | input/output pagmo::population to be evolved. |
Implements pagmo::algorithm::base.
|
protectedvirtual |
Extra human readable algorithm info.
Will return a formatted string displaying the parameters of the algorithm.
Reimplemented from pagmo::algorithm::base.
 1.8.9.1
1.8.9.1